Investigating Skewness to Understand Gene Expression Heterogeneity in Large Patient Cohorts
Abstract
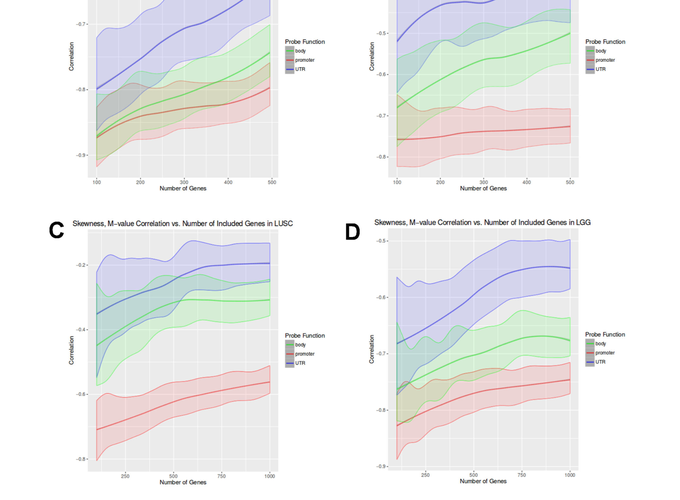
Skewness is an under-utilized statistical measure that captures the degree of asymmetry in the distribution of any dataset. This study applied a new metric based on skewness to identify regulators or genes that have outlier expression in large patient cohorts. We investigated whether specific patterns of skewed expression were related to the enrichment of biological pathways or genomic properties like DNA methylation status. Our study used publicly available datasets that were generated using both RNA-sequencing and microarray technology platforms. For comparison, the datasets selected for this study also included different samples derived from control donors and cancer patients. When comparing the shift in expression skewness between cancer and control datasets, we observed an enrichment of pathways related to immune function that reflect increases towards positive skewness in the cancer relative to control datasets. Significant correlation was also detected between expression skewness and differential DNA methylation occurring in the promotor regions for four TCGA cancer cohorts. Our results indicate the expression skewness can reveal new insights into transcription based on outlier and asymmetrical behaviour in large patient cohorts.